Complex DNA replication machinery couldn't have evolved during one bacterial life cycle
Binary fission
Asexual reproduction by a separation of the body into two new bodies. In the process of binary fission, an organism duplicates its genetic material, or deoxyribonucleic acid (DNA), and then divides into two parts (cytokinesis), with each new organism receiving one copy of DNA. (britannica.com)
How long does a bacterium live?
So, if someone claims evolution took place within first assumed bacteria (or other prokaryotes), then he/she believes that one bacterium evolved the complex DNA replication machinery just during ONE BACTERIAL LIFE CYCLE. In 12 hours or so!! And now we have to have a look at this complex DNA replication mechanism:
https://www.onlinebiologynotes.com/dna-replication/
https://www.onlinebiologynotes.com/dna-replication/
DNA replication in prokaryotes
1. Initiation:
DNA replication begins from origin. In E coli, replication origin is called OriC which consists of 245 base pair and contains DNA sequences that are highly conserved among bacterial replication origin. Two types of conserved sequences are found at OriC, three repeats of 13 bp (GATRCTNTTNTTTT) and four/five repeats of 9 bp (TTATCCACA) called 13 mer and 9 mer respectively.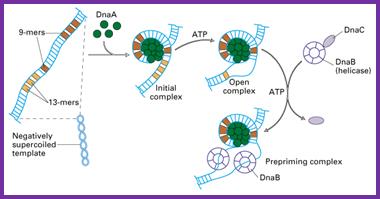
- About 20 molecules of Dna A proteins binds with 9 mer repeats along with ATP which causes DNA to wraps around dnaA protein forming initial complex. The dna A protein and ATP trigger the opening of 13 mer repeats froming open complex.
- Two copies of dnaB proteins (helicase) binds to 13 mer repeats. This binding is facilitated by another molecule called dnaC. The dnaB-dnaC interaction causes dnaB ring to open which binds with each of the DNA strand. The hydrolysis of bound ATP release dnaC leaving the dnaB bound to the DNA strand.
- The binding of helicase is key step in replication initiation. dnaB migrates along the single stranded DNA in 5’-3’ direction causing unwinding of the DNA.
- The activity of helicase causes the topological stress to the unwinded strand forming supercoiled DNA. This stress is relieved by the DNA topoisomerase (DNA gyrase) by negative supercoiling. Similarly, single stranded binding protein binds to th separated strand and prevents reannaeling of separated strand and stabilize the strand.
- The DNA polymerase cannot initiate DNA replication. So, at first primase synthesize 10±1 nucleotide (RNA in nature) along the 5’-3’ direction. In case of E.coli primer synthesized by primase starts with ppp-AG-nucleotide. Primer is closely associated with dnaB helicase so that it is positioned to make RNA primer as ssDNA of lagging strand.
2. Elongation: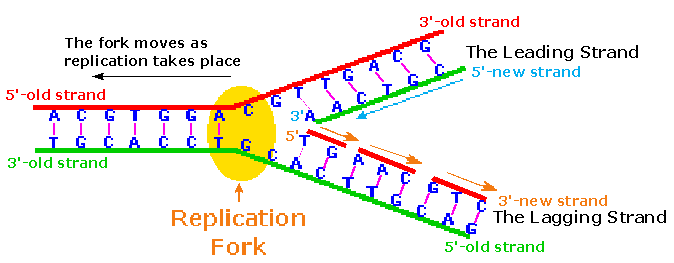
i. Leading strand synthesis:
- Leading strand synthesis is more a straight forward process which begins with the synthesis of RNA primer by primase at replication origin.
- DNA polymerase III then adds the nucleotides at 3’end. The leading strand synthesis then proceed continuously keeping pace with unwinding of replication fork until it encounter the termination sequences.
ii. Lagging strand synthesis:
- The lagging strand synthesized in short fragments called Okazaki fragments. At first RNA primer is synthesized by primase and as in leading strand DNA polymerase III binds to RNA primer and adds dNTPS.
- On this level the synthesis of each okazaki fragments seems straight forward but the reality is quite complex.
Mechanism of Lagging strand synthesis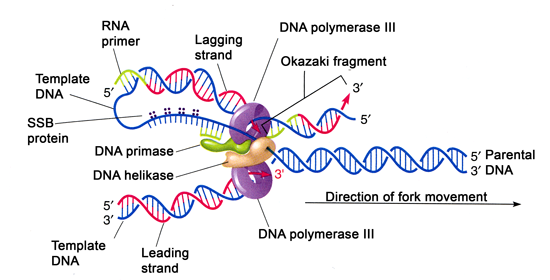
- The complexicity lies in the co-ordination of leading and lagging strand synthesis. Both the strand are synthesized by a single DNA polymerase III dimer which accomplished the looping of template DNA of lagging strand synthesizing Okazaki fragments.
- Helicase (dnaB) and primase (dnaG) constitute a functional unit within replication complex called primosome.
- DNA pol III use one set of core sub unit (Core polymerase) to synthesize leading strand and other set of core sub unit to synthesize lagging strand.
- In elongation steps, helicasein front of primaseand pol III, unwind the DNA at the replication fork and travel along lagging strand template along 5’-3’ direction.
- DnaG primase occasionally associated with dnaB helicase synthesizes short RNA primer. A new B-sliding clamp is then positioned at the primer by B-clamp loading complex of DNA pol III.
- When the Okazaki fragments synthesis is completed, the replication halted and the core sub unit dissociates from their sliding clamps and associates with new clamp. This initiates the synthesis of new Okazaki fragments.
- Both leading and lagging strand are synthesized co-ordinately and simultaneously by a complex protein moving in 5’-3’ direction. In this way both leading and lagging strand can be replicated at same time by a complex protein that move in same direction.
- Every so often the lagging strands must dissociates from the replicosome and reposition itself so that replication can continue.
- Lagging strand synthesis is not completes until the RNA primer has been removed and the gap between adjacent Okazaki fragments are sealed. The RNA primer are removed by exonuclease activity (5’-3’) of DNA pol-I and replaced by DNA. The gap is then sealed by DNA ligase using NAD as co-factor.
Termination:
- Evantually the two replication fork of circular E. coli chromosome meet at termination recognizing sequences (ter).
- The Ter sequence of 23 bp are arranged on the chromosome to create trap that the replication fork can enter but cannot leave. Ter sequences function as binding site for TUS protein.
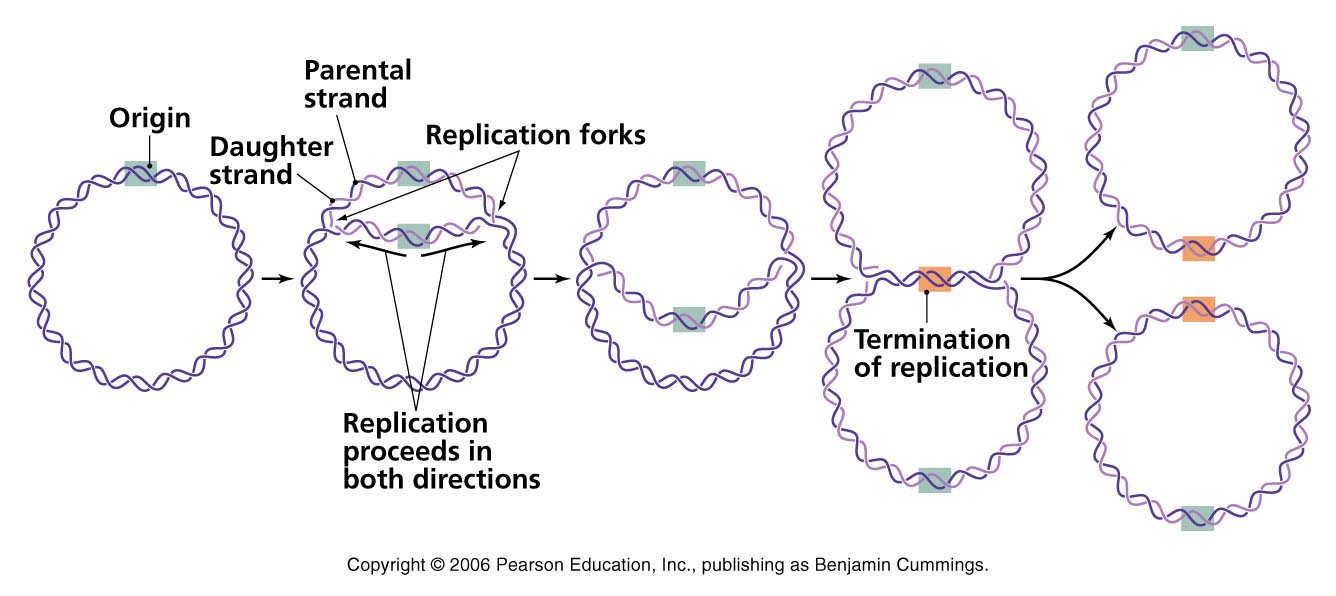
- Ter-TUS complex can arrest the replication fork from only one direction. Ter-TUS complex encounter first with either of the replication fork and halt it. The other opposing replication fork halted when it collide with the first one. This seems the Ter-TUS sequences is not essential for termination but it may prevents over replication by one fork if other is delayed or halted by a damage or some obstacle.
- When either of the fork encounter Ter-TUS complex, replication halted.
- Final few hundred bases of DNA between these large protein complexes are replicated by not yet known mechanism forming two interlinked (cataneted) chromosome.
- In E. coli DNA topoisomerase IV (type II) cut the two strand of one circular DNA and segrate each of the circular DNA and finally join the strand. The DNA finally transfer to two daughter cell.
How did the first assumed unicellular organism evolve this complex DNA replication machinery just during its short lifespan? Remember that reproduction and DNA replication are not possible if just one element is missing in this complex machinery.